The Subread aligner: fast, accurate and scalable read mapping by seed-and-vote
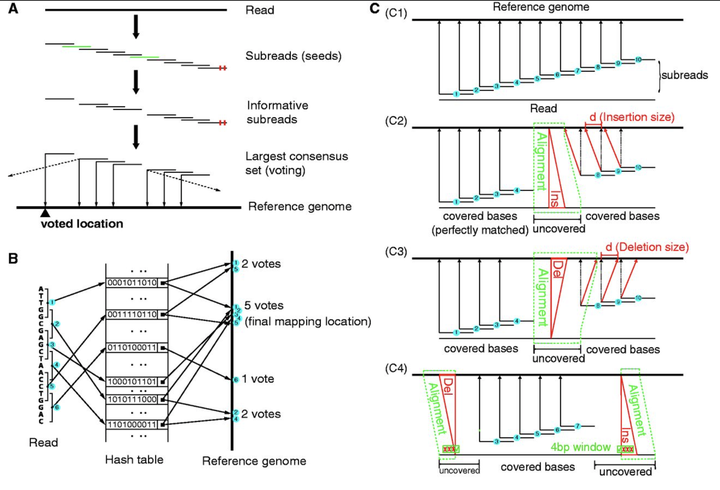 Image credit:
Nucleic Acids Research
Image credit:
Nucleic Acids Research
Yang Liao, Gordon K. Smyth, Wei Shi, The Subread aligner: fast, accurate and scalable read mapping by seed-and-vote, Nucleic Acids Research, Volume 41, Issue 10, 1 May 2013, Page e108, https://doi.org/10.1093/nar/gkt214
Read alignment is an ongoing challenge for the analysis of data from sequencing technologies. This article proposes an elegantly simple multi-seed strategy, called seed-and-vote, for mapping reads to a reference genome. The new strategy chooses the mapped genomic location for the read directly from the seeds. It uses a relatively large number of short seeds (called subreads) extracted from each read and allows all the seeds to vote on the optimal location. When the read length is <160 bp, overlapping subreads are used. More conventional alignment algorithms are then used to fill in detailed mismatch and indel information between the subreads that make up the winning voting block. The strategy is fast because the overall genomic location has already been chosen before the detailed alignment is done. It is sensitive because no individual subread is required to map exactly, nor are individual subreads constrained to map close by other subreads. It is accurate because the final location must be supported by several different subreads. The strategy extends easily to find exon junctions, by locating reads that contain sets of subreads mapping to different exons of the same gene. It scales up efficiently for longer reads.
Hola chicos! Les dejo el paper del software que usamos para el alineamiento de las lecturas de RNA-Seq en el TP3. Además acá abajo tienen el link a otro paper del año pasado en el que presentaron una versión de Subread para R (Rsubread). La principal ventaja de correrlo en R es que se mantiene todo el flujo de trabajo dentro del mismo environment, ya que la mayor parte de los análisis downstream que se realizan luego de llegar a la matriz de expresión (o counts) se hace con paquetes en R.
Yang Liao, Gordon K Smyth, Wei Shi, The R package Rsubread is easier, faster, cheaper and better for alignment and quantification of RNA sequencing reads, Nucleic Acids Research, Volume 47, Issue 8, 07 May 2019, Page e47, https://doi.org/10.1093/nar/gkz114